Bayesian Linear Regression with SGD, Adam and NUTS in PyTorch
Published:
PyTorch has gained great popularity among industrial and scientific projects, and it provides a backend for many other packages or modules. It is also accompanied with very good documentation, tutorials, and conferences. This blog attempts to use PyTorch to fit a simple linear regression via three optimisation algorithms:
Stochastic Gradient Descent (SGD)
Adam
No-U-Turn Sampler (NUTS)
We will start with some simulated data, given certain parameters, such as weights, bias and sigma. The linear regression can be expressed by the following equation:
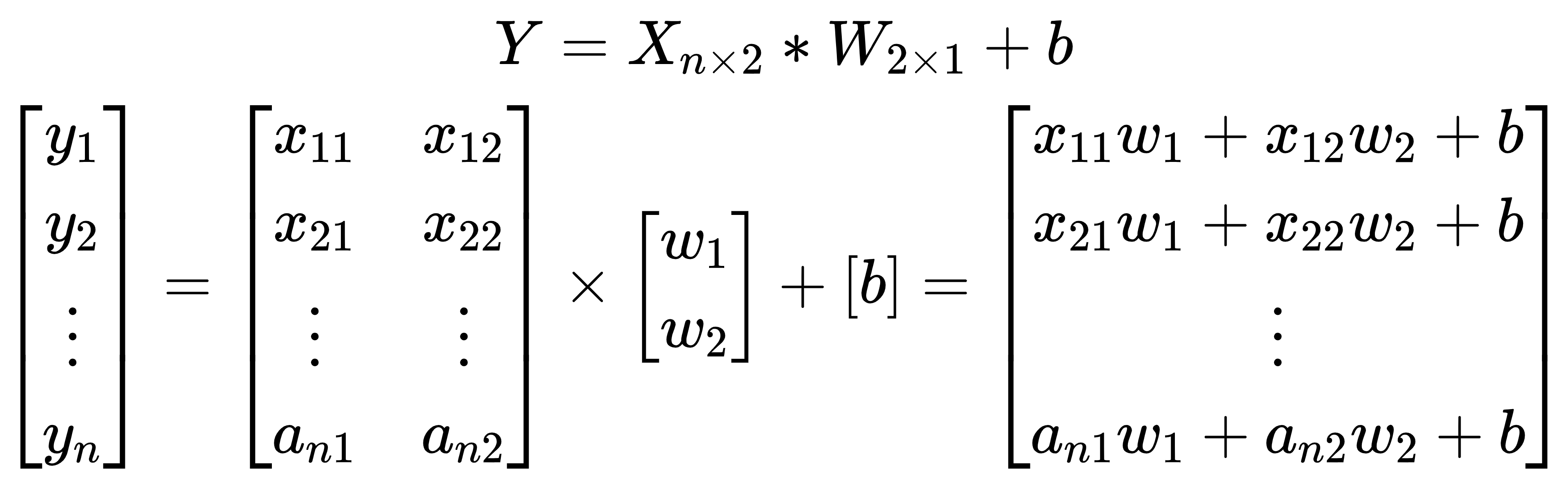
where the observed dependent variable Y is a linear combination of data (X) times weights (W), and add the bias (b). This is essentially the same as the *nn.Linear* class in PyTorch.
1. simulate data
We need to load the dependent modules, such as torch, jax, and numpyro.
from __future__ import print_function
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.distributions.normal import Normal
from torch.distributions.uniform import Uniform
from torch.distributions.exponential import Exponential
import jax
from jax import random
from jax import grad, jit
import jax.numpy as np # using jax.numpy instead
import numpyro
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS
Let’s simulate the data with pre-defined parameters. Here we have 50 observations, 2 weights, 1 bias, and 1 sigma.
N = 50; J = 2
X = random.normal(random.PRNGKey(seed = 123), (N, J))
weight = np.array([1.5, -2.8])
error = random.normal(random.PRNGKey(234), (N, )) # standard Normal
b = 10.5
y_obs = X @ weight + b + error
y = y_obs.reshape((N, 1))
X = jax.device_get(X) # convert jax array into numpy array
y = jax.device_get(y) # convert jax array into numpy array
x_data = Variable(torch.from_numpy(X), requires_grad=True)
y_data = Variable(torch.from_numpy(y), requires_grad=True)
2. Models
(1) SGD
The first optimisation method is SDG, and it updates the parameters in the directions of gradients of each parameters so as to minimise the loss function, such as nn.MSELoss in this case. The chunk below defines your own class of linear model.
class lm_model(nn.Module):
def __init__(self, in_features, out_features, bias=True):
super().__init__()
self.in_features = in_features
self.out_features = out_features
self.bias = bias
self.weight = torch.nn.Parameter(torch.randn(out_features, in_features))
self.bias = torch.nn.Parameter(torch.randn(out_features))
def forward(self, input):
x, y = input.shape
if y != self.in_features:
print(f'Wrong Input Features. Pls use tensor with {self.in_features} Input Features')
return 0
output = input.matmul(self.weight.t())
if self.bias is not None:
output += self.bias
return output
We can run the analysis:
model = lm_model()
criterion = nn.MSELoss(reduction="sum")
optimizer = torch.optim.SGD(model.parameters(), lr = 0.01)
for epoch in range(8000):
y_predict = model(x_data)
loss = criterion(y_predict, y_data)
if (epoch + 1) % 2000 == 0 or epoch % 2000 == 0:
print(epoch)
print("Estimated weights: ", model.linear.weight.data)
print("Estimated bias: ", model.linear.bias.data.item())
print("Estimated loss: ", loss.data.item())
print("====================")
optimizer.zero_grad()
loss.backward()
optimizer.step()
(2) Adam
We can also define your model with a likelihood-based method, and search for the parameters to maximise the likelihood function via Adam. In other words, with the data observed (x and y), we are looking for a set of parameters, alpha, beta and sigma, to maximise the likelihood function of a Normal distribution. \(P(y | x, \alpha, \beta, \sigma)= \mathbf{N}\left(y | \alpha x + \beta , \sigma \right)\)
class lm_model_lik(nn.Module):
def __init__(self):
super().__init__()
ws = Uniform(-10, 10).sample((3, 1))
self.weights = torch.nn.Parameter(ws)
self.sigma = torch.nn.Parameter(Uniform(0, 2).sample())
def forward(self, input, output):
prior_weights = Normal(0, 10).log_prob(self.weights).sum()
prior_sigma = Exponential(2.0).log_prob(self.sigma)
i, _= input.shape
y_hat = input @ self.weights[0:2] + self.weights[2:3]
y_hat = y_hat.view(i, -1)
lik = Normal(y_hat, self.sigma).log_prob(output).sum()
LL = lik + prior_weights + prior_sigma
return -LL
We can run the analysis with the Adam sampler:
model = lm_model_lik()
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
for epoch in range(10000):
neg_log_lik = model(x_data, y_data)
if (epoch + 1) % 1000 == 0 or epoch % 1000 == 0:
print(epoch)
print("Estimated beta1: ", model.weights.data.view(3))
print("Estimated sigma: ", model.sigma.item())
print("Estimated neg logLik: ", neg_log_lik.item())
print("====================")
optimizer.zero_grad()
neg_log_lik.backward()
optimizer.step()
(3) NUTS
It is also possible to use the NUTS to estimate the parameters in the linear model. This can be either likelihood-based (see below) or distance-based with a loss function (akin to Approximate Bayesian Computation).
def model(X, y=None):
ndims = np.shape(X)[-1]
ws = numpyro.sample('betas', dist.Normal(0.0,10*np.ones(ndims)))
b = numpyro.sample('b', dist.Normal(0.0, 10.0))
sigma = numpyro.sample('sigma', dist.Uniform(0.0, 10.0))
mu = X @ ws + b
return numpyro.sample("y", dist.Normal(mu, sigma), obs = y)
The model is fitted by the NUTS sampler.
X = jax.device_put(X) # convert numpy array into jax array
y = jax.device_put(y) # convert numpy array into jax array
nuts_kernel = NUTS(model)
num_warmup, num_samples = 500, 500
mcmc = MCMC(nuts_kernel, num_warmup, num_samples)
mcmc.run(random.PRNGKey(0), X, y = y_obs)
mcmc.get_samples()
Useful links: