Checkpoints in stan via chkptstanr
Published:
In this blog, I will introduce a new R package chkptstanr, which allows checkpoints in stan. This feature is useful when you want to cache the model outputs by certain time steps or resume a long run at specific points due to an interruption. In general, this package can be considered as a wrapper for cmdstan, and thus you need to download and install cmdstan on your machine. Here are the step-by-step instruction on how you can deploy the environment and run the stan model via chkptstanr.
1. Install cmdstan
The cmdstan is the shell interface to stan and can be integrated well with the command-line interface. It also supports GPU computation via OpenCL and multi-threading (see the latest version of cmdstan user’s guide). You can install the cmdstan by the following commands.
# download cmdstan-2.29.2
wget https://github.com/stan-dev/cmdstan/releases/download/v2.29.2/cmdstan-2.29.2.tar.gz
tar -xf cmdstan-2.29.2.tar.gz
cd cmdstan-2.29.2
make build
If you are interested in setting up cmdstan with GPU support, I am including my configuration for the Snowy cluster.
# load the necessary packages
module use /sw/EasyBuild/snowy/modules/all/
module load intelcuda/2019b
module load gcc/9.3.0
module load R/4.1.1
module load R_packages/4.1.1
# set the local configuration
cd cmdstan-2.29.2
echo "STAN_OPENCL=TRUE" > make/local
make build
Let’s check the configuration by running an example stan model (bernoulli) inside the cmdstan folder.
cd cmdstan-2.29.2
ls example/bernoulli
# bernoulli.data.R bernoulli.data.json bernoulli.stan
# Note: compile the model without the suffix of .stan
make example/bernoulli/bernoulli
examples/bernoulli/bernoulli sample data \
file=examples/bernoulli/bernoulli.data.json output file=output.csv
2. Run stan model via chkptstanr
Now we have cmdstan installed on your computer and we can try to run a stan model via chkptstanr. Before that we need to load the necessary packages and specify the cmdstan path. Here I am using a version from github.
library(cmdstanr)
# library(devtools)
# install_github("donaldRwilliams/chkptstanr")
library(chkptstanr)
library(bayesplot)
set_cmdstan_path("../cmdstan-2.29.2")
(1) Model
For simplicity, I am using a stan model from the chkptstanr package by adjusting the model with the new array syntax, and you can of course try your own stan model.
stan_code <- "
data {
int<lower=0> n;
array[n] real y;
array[n] real<lower=0> sigma;
}
parameters {
real mu;
real<lower=0> tau;
vector[n] eta;
}
transformed parameters {
vector[n] theta;
theta = mu + tau * eta;
}
model {
target += normal_lpdf(eta | 0, 1);
target += normal_lpdf(y | theta, sigma);
}
"
(2) Data
stan_data <- list(n = 8,
y = c(28, 8, -3, 7, -1, 1, 18, 12),
sigma = c(15, 10, 16, 11, 9, 11, 10, 18))
(3) Run the analysis
It is worth noting that when you first run the model, you often need to create a folder via create_folder. But if you want to resume a previous run, you just need to specify the path to previously created folder. To achieve this, we include a conditional statement to check the existence of the folder.
# important: check for the existence of folder
name_folder = "stan_chkpt_m1"
if(dir.exists(name_folder)){
path = paste0("./", name_folder)
}else{
path = create_folder(folder_name = name_folder)
}
# important: specify the stan_code_path
stan_code_path = paste0(path, "/stan_model/model.stan")
fit_m1 <- chkpt_stan(model_code = stan_code, data = stan_data,
iter_warmup = 1000, iter_sampling = 2000,
iter_per_chkpt = 250, path = path)
You can directly stop the running model or specify a longer sampling iteration. Normally, chkpt_stan will resume the model from the cached checkpoints, no matter whether they occur at the warm-up or sampling stage. Another advantage is that if the checkpointing is completed, the model won’t be executed even though you run the same command once again. If you really want to rerun the model, you can delete the cached folder by using the unlink or system function.
# remove the cached folder
unlink(path, recursive = T)
# system(command = paste0("rm -r ", path))
It is worth noting that the current version of chkptstanr does not allow you to resize the warm-up iterations or change the iteration per checkpoint between two runs, since they may cause some difficulties to combine different checkpoint draws.
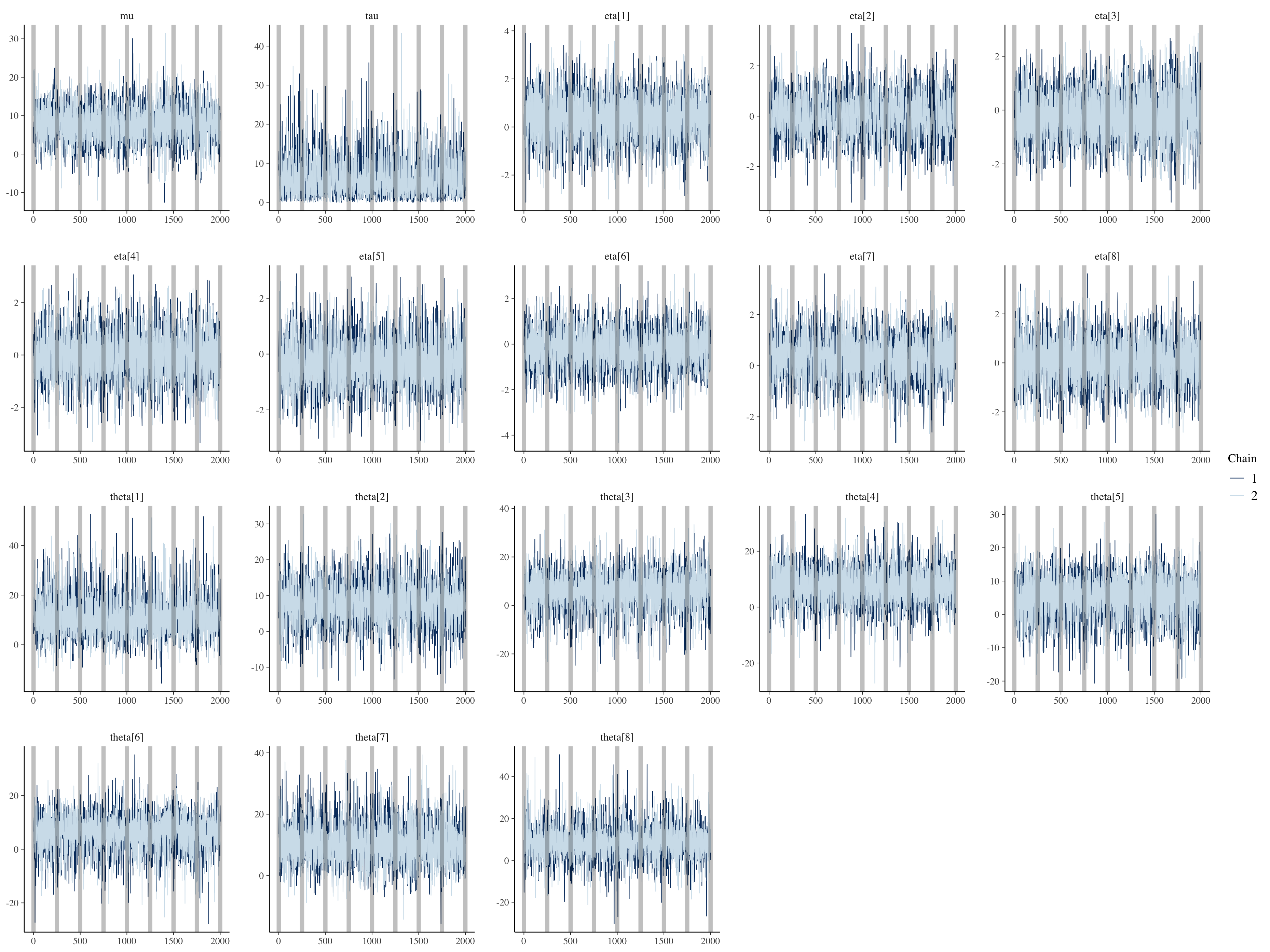
(4) Visualization
draws <- combine_chkpt_draws(fit_m1)
bayesplot::mcmc_trace(draws, regex_pars = c("mu", "tau", "eta")) +
geom_vline(xintercept = seq(0, 2000, 250), alpha = 0.25, size = 2)
Useful links: